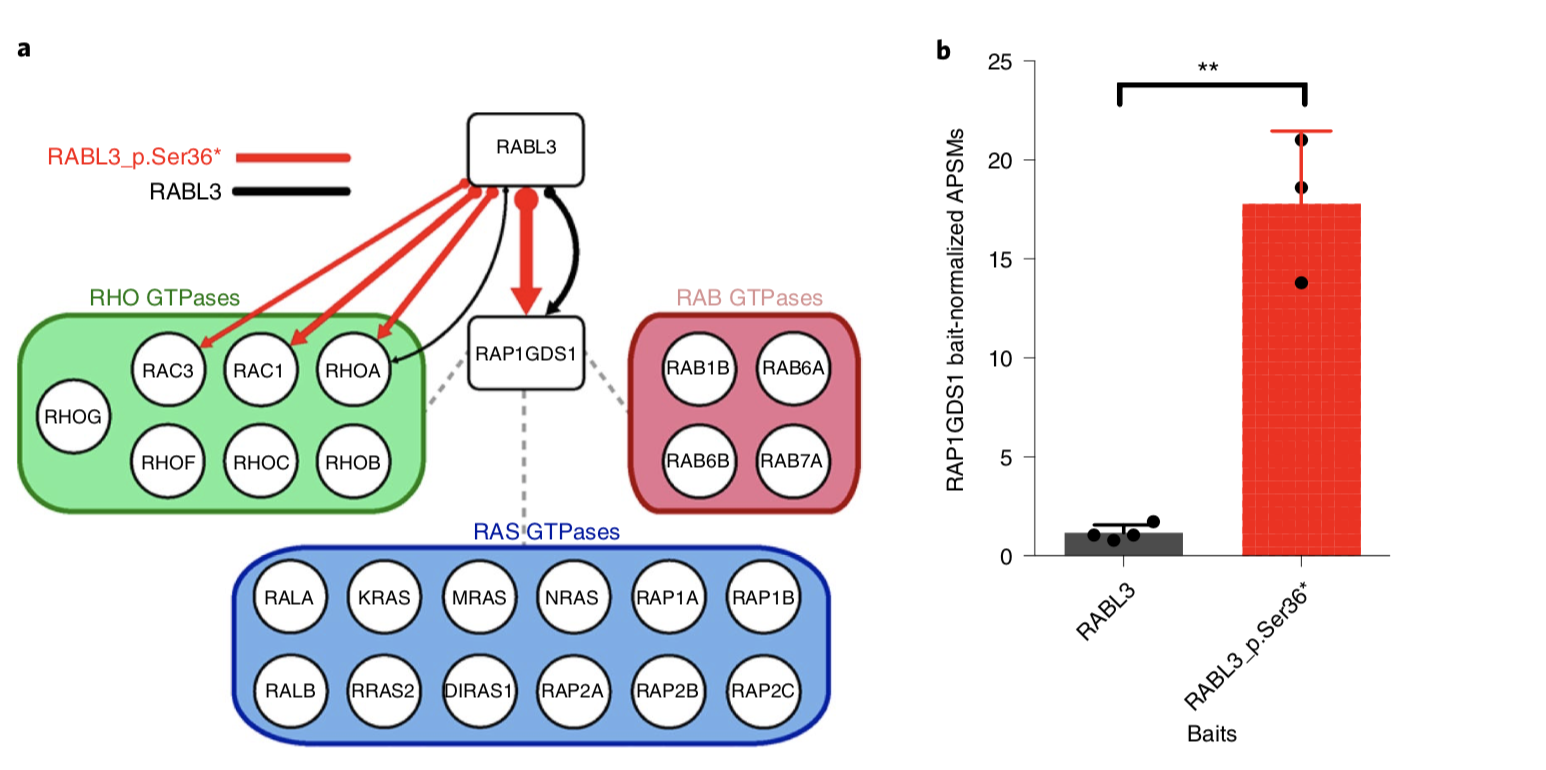
| Interaction proteomics of RABL3 wild-type and mutant: a Integrated interaction map of the RABL3 and RABL3_p.Ser36* network showing high-confidence candidate interacting proteins. Thickness of RABL3 (black) and RABL3_p.Ser36* (red) arrows denotes NWD scores. RAP1GDS1 interactors identified by AP–MS with RAP1GDS1-tagged bait are grouped by protein family for clarity and denoted by dashed gray lines. b, RAP1GDS1 bait-normalized APSMs for RABL3 versus RABL3_p.Ser36* AP–MS. Graph represents mean ± s.d., independent biological replicates N = 3 for RABL3_p.Ser36*, N = 4 for RABL3. **P = 0.0014 by unpaired two-tailed t-test (t = 7.813, d.f. = 4) | Sahar Nissim from Wolfram Goessling's Lab leads a study identifying a mutation in RABL3 that is associated with hereditary pancreatic ductal adenocarcinoma. Joe initially helped Sahar to identify the molecular mechanism of RABL3 by performing interaction proteomics of RABL3 and RABL3 mutants. To the left is some of the data from the proteomics. Great work from Sahar - this study now nominates RABL3 as a target for genetic testing in cancer families |
|
0 Comments
|
Archives
September 2022
Categories |



 RSS Feed
RSS Feed